Home
 |
||||||
|
|
|||||
Research
Our research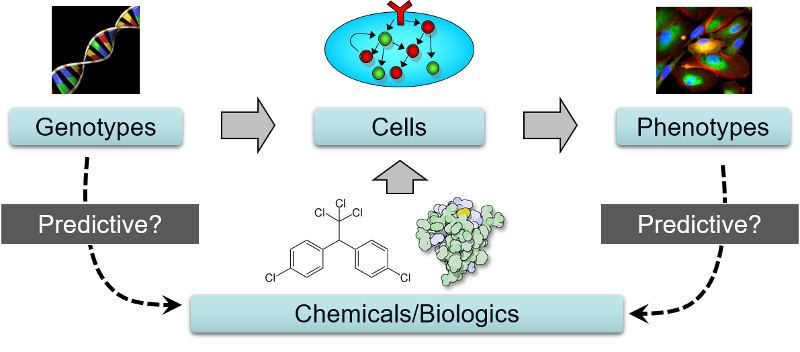 Toxicodynamics of xenobiotics
Many xenobiotics have unknown and/or non-specific intracellular targets. To study the toxicodynamics of these chemicals, unbiased approaches that do not require prior information about the targets or mechanisms of the chemicals are required. Our goal is to elucidate the MoAs of xenobiotics in major target cell types using advanced phenotypic, signaling, and genomic profiling methods.
Signaling responses: Besides cell injury, toxic xenobiotics may also induce signaling or inflammatory responses in their targeted cell types. We have developed a rapid, signaling-based cytotoxicity assay that may be used to predict cellular sensitivity to a cytotoxic agent, or identify co-treatments that may sensitize or desensitize cells to the agent (Loo et al., 2017b). We have also developed a predictive nephrotoxicity assay based on the RNA expression levels of two pro-inflammatory cytokines, namely interleukin (IL)-6 and -8 (Kandasamy et at., 2015; Su et al., 2014), and tested it on both primary human PTCs and induced pluripotent stem cells (iPSC)-derived PTC-like cells. These results suggest that inflammation is a general response of PTCs to PTC-toxic compounds. Transcriptomic responses: The expressions of genes involved in key toxicity responses may be up- or down-regulated in response to toxic xenobiotics. Recent advances in transcriptomics technologies have enabled us to quantify these changes at the genome-wide level. In collaboration with Dr. Hoon from MEL, we are developing high-throughput transcriptomic methods to study concentration-dependent changes in key toxicity pathways. Toxicity Mode-of-Action Discovery (ToxMAD) Platform: Together with four other research institutes in A*STAR, we are using various new molecular and phenotypic profiling technologies developed in A*STAR to elucidate the protein targets and MoAs of xenobiotics with high human exposure or safety concerns. Our focus is to study chemical analogs with related structures but differential cellular effects, and develop fit-for-purpose assays that will be used by regulatory agencies and industrial research laboratories to assess chemical safety. 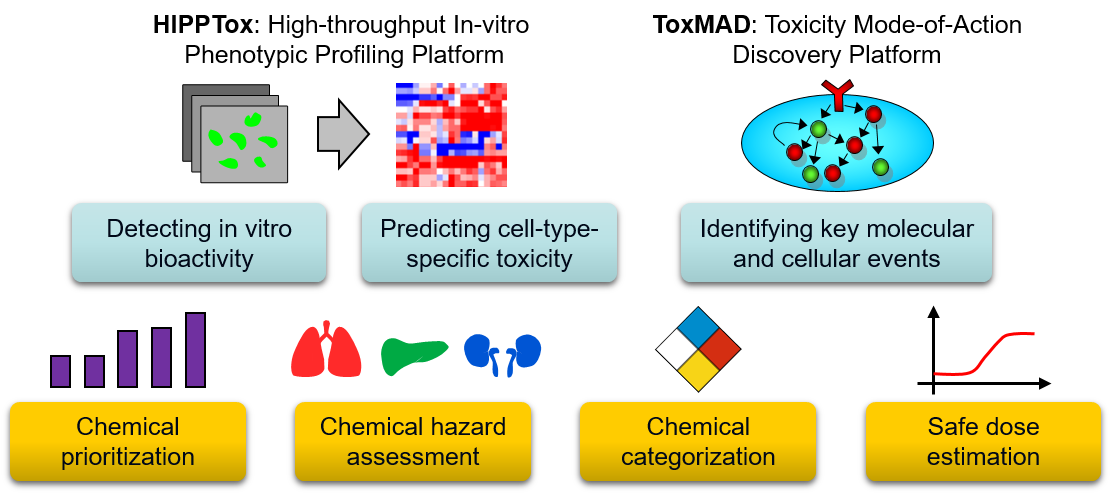 Pulmonary effects of xenobiotics
Human lungs are exposed to inhaled or blood-borne soluble xenobiotics that may originate from the environment, food, consumer products, and/or pharmaceuticals. We are broadly interested in the understanding the biological targets and pathways affected by these chemicals in the lung cells. In vitro toxicity models: We have recently developed a high-throughput and predictive in vitro pulmonary toxicity assay (Lee et al., 2018). We found that the resulting assay based on two phenotypic features of a human bronchial epithelial cell line, BEAS-2B, can accurately classify 33 reference chemicals with human pulmonotoxicity information (88.8% balance accuracy, 84.6% sensitivity, and 93.0% specificity). In comparison, the predictivity of a standard cell-viability assay on the same set of chemicals is much lower (77.1% balanced accuracy, 84.6% sensitivity, and 69.5% specificity). We also used the assay to evaluate 17 additional test chemicals with unknown/unclear human pulmonotoxicity, and experimentally confirmed that many of the pulmonotoxic reference and predicted-positive test chemicals induce DNA strand breaks and/or activation of the DNA-damage response (DDR) pathway. 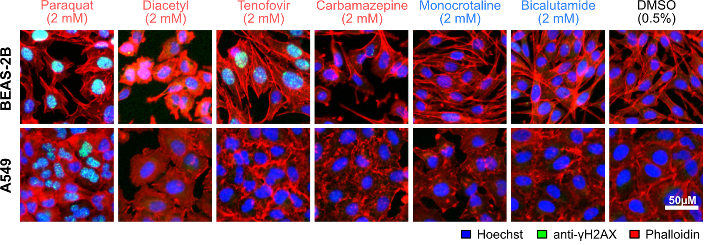 Xenobiotic metabolism: In the lungs, bronchial and alveolar epithelial cells are major sites of xenobiotic metabolism, and thus are susceptible to the toxicity induced by xenobiotics that interfere with this process. In collaboration with Dr. Hao Fan from BII, we are studying the mechanisms of xenobiotics that can inhibit Cytochrome P450 family 1 member A1 (CYP1A1), a main extra-hepatic phase I metabolism enzyme highly expressed in the lungs and placenta. We have developed molecular docking models that can be used to predict potential CYP1A1 inhibitors. Phenotypic profiling and computational biology
To extract biological information from the large amount of collected data, new and better methods and tools for image and data analysis are required. Most of our projects are based on the HIPPTox Platform and the cellXpress software developed by us. Our group also develops new methodologies for concentration response modeling, artificial intelligence, and assay automation.
Concentration response modeling: A concentration response curve (CRC) is commonly used to model the relationship between the concentration and effect of a perturbagen. However, for automated perturbagen classification based on quantitative phenotypic features from HCI, it is unclear if commonly used CRC metrics, such as the "half-maximal effective concentration" (EC50) that reports perturbagen potency, are still optimal. We have performed a systematic study on the performances of different CRC metrics in classifying four HCI datasets that consist of phenotypic features from different cell and feature types. Our results suggest that efficacy metrics, especially at higher concentration values, are more likely to provide the most useful information for perturbagen classification. Therefore, HCI experiments should include measurements at high perturbagen concentrations, and efficacy metrics should always be analyzed when building supervised classifiers based on phenotypic features. |
Publications
Publications |
|
| 29. | (2023) Global Epidemiology and Genetics of Hepatocellular Carcinoma Ming Ren Toh, Evelyn Yi Ting Wong, Sunny Hei Wong, Alvin Wei Tian Ng, Lit-Hsin Loo, Pierce Kah-Hoe Chow, Joanne Yuen Yie Ngeow. Gastroenterology, in press [Link]. |
| 28. | (2022) Choice of PD-L1 immunohistochemistry assay influences clinical eligibility for gastric cancer immunotherapy Joe Yeong, Huey Yew Jeffrey Lum, Chong Boon Teo, Benjamin Kye Jyn Tan, Yiong Huak Chan, Ryan Yong Kiat Tay, Joan Rou-En Choo, Anand D Jeyasekharan, Qing Hao Miow, Lit-Hsin Loo, Wei Peng Yong, Raghav Sundar. Gastric Cancer, 25:741–750 [Link]. |
| 27. | (2021) 627 ImmunoAtlas: an online public portal for sharing, visualizing, and referencing multiplex immunohistochemistry/immunofluorescence (mIHC/IF) images and results for immuno-oncology Jia Ying Joey Lee, Joe Yeong, Li Wen Justina Nadia Lee, Lit-Hsin Loo and Jiahui Dong. J Immunother Cancer, 9(Suppl 2):A657 [Link]. |
| 26. | (2021) Group VIII Metal Carbonyl Cluster-Boronic Acid Conjugates: Cytotoxicity and Mode of Action Studies Jia Wen Kong, Zhiyong Lam, Kiat Hwa Chan, Rakesh Ganguly, Jia-Ying Joey Lee, Lit-Hsin Loo, Richard D. Webster, Zhen Xuan Wong, and Weng Kee Leong. ACS Omega, 6(43):29045–29053 [Link]. |
| 25. | (2021) Leveraging advances in immunopathology and artificial intelligence to analyze in vitro tumor models in composition and space Matthew Leong Tze Ker, Lo Wen Shern, Zen Lee Wei Ena, Benedict Tan, Lee Xing Zhao, Lee Li Wen Justina Nadia, Jia-Ying Joey Lee, Nivedita Suresh, Lit-Hsin Loo, Evan Szu, Joe Yeong. Advanced Drug Delivery Reviews, 177:113959 [Link]. |
| 24. | (2021) Structure-based virtual screening of CYP1A1 inhibitors: towards rapid tier-one assessment of potential developmental toxicants Janice Jia Ni Goh, Julian Behn, Cheng-Shoong Chong, Guorui Zhong, Sebastian Maurer-Stroh, Hao Fan & Lit-Hsin Loo. Archives of Toxicology, 95, 3031-3048 [Link]. |
| 23. | (2021) Virtual screening of potentially endocrine-disrupting chemicals against nuclear receptors and its application to identify PPARγ-bound fatty acids Chaitanya K. Jaladanki, Yang He, Li Na Zhao, Sebastian Maurer-Stroh, Lit-Hsin Loo, Haiwei Song, and Hao Fan. Archives of Toxicology, 95:355-374 [Link]. |
| 22. | (2020) Optimum concentration–response curve metrics for supervised selection of discriminative cellular phenotypic endpoints for chemical hazard assessment James Miller, and Lit-Hsin Loo. Archives of Toxicology, 94:2951-2964 [Link]. |
| 21. | (2020) Predicting direct hepatocyte toxicity in humans by combining high-throughput imaging of HepaRG cells and machine learning-based phenotypic profiling Faezah Hussain, Sreetama Basu, Javen Jun Hao Heng, Lit-Hsin Loo, and Daniele Zink. Archives of Toxicology, 94:2749-2767 [Link]. |
| 20. | (2020) A case study with triazole fungicides to explore practical application of next generation hazard assessment methods for human health. Leo Van Der Ven, Emiel Rorije, Corinne Sprong, Daniele Zink, Remco Derr, Giel Hendriks, Lit-Hsin Loo, and Mirjam Luijten. Chemical Research in Toxicology, 33(3):834–848 [Link]. |
| 19. | (2020) Utility of in vitro bioactivity as a lower bound estimate of in vivo adverse effect levels and in risk-based prioritization. Katie Paul Friedman, Matthew Gagne, Lit-Hsin Loo, Panagiotis Karamertzanis, Tatiana Netzeva, Tomasz Sobanski, Jill Franzosa, Ann Richard, Ryan Lougee, Andrea Gissi, Jia-Ying Joey Lee, Michelle Angrish, Jean-Lou Dorne, Stiven Foster, Kathleen Raffaele, Tina Bahadori, Maureen Gwinn, Jason Lambert, Maurice Whelan, Mike Rasenberg, Tara Barton-Maclaren, Russell S Thomas. Toxicological Sciences, 173(1):202-225 [Link]. |
| 18. | (2019) PI3K catalytic subunits α and β modulate cell death and IL-6 secretion induced by talc particles in human lung carcinoma cells. Nicola Michelle Bougen-Zhukov, Yin Yeng Lee, Jia-Ying Joey Lee, Pyng Lee, and Lit-Hsin Loo. American Journal of Respiratory Cell and Molecular Biology, [Link]. |
| 17. | (2018) Emerging technologies for food and drug safety. William Slikker Jr., Thalita Antonyde Souza Lima, Davide Archella, Jarbas Barbosa de Silva Junior, Tara Barton-Maclaren, Li Bo, Danitza Buvinich, Qasim Chaudhry, Peiying Chuang, Hubert Deluyker, Gary Domselaar, Meiruze Freitas, Barry Hardy, Hans-Georg Eichler, Marta Hugas, Kenneth Lee, Chia-Ding Liao, Lit-Hsin Loo, Haruhiro Okuda, Orish Ebere Orisakwe, Anil Patri, Carl Sactitono, Leming Shi, Primal Silva, Frank Sistare, Shraddha Thakkar, Weida Tong, Mary Lou Valdez, Maurice Whelan, and Anna Zhao-Wong. Regulatory Toxicology and Pharmacology, 98:115-128. [Link] |
| 16. | (2018) Building predictive in vitro pulmonary toxicity assays using high-throughput imaging and artificial intelligence. Jia-Ying Joey Lee, James Alastair Miller, Sreetama Basu, Ting-Zhen Vanessa Kee, and Lit-Hsin Loo. Archives of Toxicology, 92(6):2055-2075. [Link] |
| 15. | (2017) High-throughput prediction of nephrotoxicity in humans. Lit-Hsin Loo, Daniele Zink. Alternatives to Laboratory Animals, 45:241-252. [PDF] |
| 14. | (2017) Early spatiotemporal-specific changes in intermediate signals are predictive of cytotoxic sensitivity to TNFa and co-treatments. Lit-Hsin Loo, Nicola Bougen-Zhukov, Wei-Ling Cecilia Tan. Scientific Reports, 7:43541. [Link] |
| 13. | (2017) Large-scale image-based screening and profiling of cellular phenotypes. Nicola Bougen-Zhukov, Sheng Yang Loh, Hwee Kuan Lee, Lit-Hsin Loo. Cytometry Part A, 91A:115-125. [Link | PDF] |
| 12. | (2016) High-throughput imaging-based nephrotoxicity prediction for xenobiotics with diverse chemical structures. Ran Su, Sijing Xiong, Daniele Zink, and Lit-Hsin Loo. Archives of Toxicology, 90:2793-2808. [Link] |
| 11. | (2015) Prediction of drug-induced nephrotoxicity and injury mechanisms with human induced pluripotent stem cell-derived cells and machine learning methods. Karthikeyan Kandasamy, Jacqueline Chuah, Ran Su, Peng Huang, Kim Guan Eng, Sijing Xiong, Yao Li, Chun Siang Chia, Lit-Hsin Loo, and Daniele Zink. Scientific Reports, 5:12337. [Link] |
| 10. | (2014) Supervised prediction of drug-induced nephrotoxicity based on interleukin-6 and -8 expression levels. Ran Su, Yao Li, Daniele Zink, and Lit-Hsin Loo. BMC Bioinformatics, 15(Suppl 16):S16. [Link] |
| 9. | (2014) Quantitative protein localization signatures reveal an association between spatial and functional divergences of proteins. Lit-Hsin Loo, Danai Laksameethanasan, Yi-Ling Tung. PLOS Computational Biology, 10(3):e1003504. [Link] |
| 8. | (2013) cellXpress: a fast and user-friendly software platform for profiling cellular phenotypes. Danai Laksameethanasan, Rui Zhen Tan, Geraldine Wei-Ling Toh and Lit-Hsin Loo. BMC Bioinformatics14(Suppl 16):S4. [Link] |
| 7. | (2009) Heterogeneity in the physiological states and pharmacological responses of differentiating 3T3-L1 preadipocytes. Lit-Hsin Loo, Hai-Jui Lin, Dinesh K. Singh, Kathleen M. Lyons, Steven J. Altschuler and Lani F. Wu. Journal of Cell Biology, Vol. 187, No. 3, 375-384. [Link | JCB In-Focus article] |
| 6. | (2009) An approach for extensibly profiling the molecular states of cellular subpopulations. Lit-Hsin Loo, Hai-Jui Lin, Robert J. Steininger III, Yanqin Wang, Lani F. Wu and Steven J. Altschuler. Nature Methods, Vol. 6, 759-765. [Link] |
| 5. | (2007) Image-based multivariate profiling of drug responses from single cells. Lit-Hsin Loo, Lani F. Wu and Steven J. Altschuler. Nature Methods, Vol. 4, 445-453. [Link] |
| 4. | (2007) New criteria for selecting differentially expressed genes. Lit-Hsin Loo, Samuel Roberts, Leonid Hrebien, and Moshe Kam. IEEE Engineering in Medicine and Biology Magazine, Vol. 26, 17-26. |
| 3. | (2002) Classification of polypeptide spectra from rat liver samples. Lit-Hsin Loo, John Quinn, James Armitage, Hayley Cordingley, Samuel Roberts, Peter J Bugelski, Leonid Hrebien, and Moshe Kam. Proceedings of the IEEE 28th Annual Northeast Bioengineering Conference, 139-140. |
| 2. | (2002) Classification of pharmaceutical toxicity by feature analysis. John Quinn, Lit-Hsin Loo, James Armitage, Hayley Cordingley, Samuel Roberts, Peter J Bugelski, Moshe Kam, and Leonid Hrebien. Proceedings of the IEEE 28th Annual Northeast Bioengineering Conference, 211-212. |
| 1. | (2002) Cooperative Multi-agent Constellation Formation Under Sensing and Communication Constraints. Lit-Hsin Loo, Erwei Lin, Moshe Kam, and Pramod Varshney. Cooperative Control and Optimization, Vol. 66, Chapter 8, 143-169. |
Our team
Current Team Members
 |
|||||||||||||||
|
|||||||||||||||
 |
LOO Lit Hsin, Ph.D.
Lit-Hsin Loo is leading a small interdisciplinary team of scientists
at BII developing in vitro and computational models for predicting the toxicity and/or targets of
chemical compounds with diverse or unknown structures. His team has developed novel image-based
phenotypic profiling methods and tools that led to the first high-throughput and predictive in
vitro platform for nephrotoxicity prediction. Before joining BII, Dr. Loo was a
postdoctoral fellow in the Bauer Center for Genomics Research at Harvard University (2005),
and then in the Department of Pharmacology at the UTSW Medical Center, USA (2005-2010).
Awards:
|
Loo Lab Alumni
Postdoctoral Fellows:
|
Research Officers/Software Engineers:
|
Interns:
|
Life in the lab
Life in the Loo Lab
 Feb. 2021.COVID hit us, but we survived!  Aug. 2019.Farewell to our intern from Canada.  Feb 2018. Our annual lab dinner. We had a fabulous 2017.  Jan 2018. Our very own Joey was getting married!  Sept 2017. We participated in the dragon boat contest.  Aug 2017. Joey, James, and Lit-Hsin attended the WC10 at Seattle. Aug 2017. The Loo Lab Band, Seattle. |
Careers
Careers @ the Loo Lab
We are always interested in postdoctoral-fellow candidates with backgrounds relevant to our work. Every year, we also take in one to two undergraduate interns to provide them with hand-on experiences in scientific research projects. Some of our interns also contributed to our published work. If you are interested to join our group, please send your CV to Dr. Lit-Hsin Loo (loolh at bii dot a-star dot edu dot sg). Specific openings in the lab are listed below:
- Postdoctoral Fellow in Computational Biology (Spatial Multi-omics for Precision Cancer Medicine) (8 Sept 2022)
- Internship in cell biology
- Internship in computational analysis of microscopy images
|
Postdoctoral Fellow in Computational Biology (Spatial Multi-omics for Precision Cancer Medicine) Description:
A postdoctoral research fellow position in the area of spatial multi-omics for precision cancer medicine is available in the Loo lab (http://web.bii.a-star.edu.sg/~loolh) at the Bioinformatics Institute (BII), A*STAR, Singapore. The group develops next-generation spatial phenotypic profiling assays, analysis methods, and machine learning models to predict patient responses to anti-cancer drugs. The group also develops and manages the HistoPath Analytics (HPA) Platform and ImmunoAtlas (https://ImmunoAtlas.org) for automated management, visualization, and analysis of large multiplex tissue images and spatial multi-omics data. Qualifications:
Application Procedures: Applicants should send
|
|
Internship in Cell Biology Description: An internship position in the area of cell biology is available in the Loo lab at the Bioinformatics Institute (BII) in Singapore. The successful candidate will study the responses of lung epithelial cells to drugs and other chemical compounds. He/she will have the opportunity to work in a highly interdisciplinary and stimulating environment, and engage in innovative cell biology research. The preferred duration of internship is three months, and the starting date is flexible. Qualifications: Candidates must have completed university course work in the areas of biological and/or chemical sciences. Previous experience in performing cell culture work and/or molecular biology experiments is desired. Application Procedures: Applicants should contact Dr. Lit-Hsin Loo (loolh at bii dot a-star dot edu dot sg) for more information. |
|
Internship in Computational Software Development Description: An internship position in the area of computational software development is available in the Loo lab at the Bioinformatics Institute (BII) in Singapore. The successful candidate will participate in the software development of image processing and machine learning tools. He/she will have the opportunity to work in a highly interdisciplinary and stimulating environment, and learn how computer science can help biologists to make biological discovery. The preferred duration of internship is three months, and the starting date is flexible. Qualifications: Candidates must have strong knowledge in C++ programming under the Linux environment, and basic knowledge of machine learning and web programming. Previous experience in bioinformatics or molecular and cell biology is desired, but not required. Candidates with interdisciplinary training in computer science and biology are especially encouraged to apply. Application Procedures: Applicants should contact Dr. Lit-Hsin Loo (loolh at bii dot a-star dot edu dot sg) for more information. |
Contact
Contact us
The Loo Lab is located at:
Bioinformatics Institute
30 Biopolis Street, #07-01 Matrix,
Singapore 138671
Email: loolh at bii dot a-star dot edu dot sg
Tel: (65) 6478 8298
Fax: (65) 6478 9048

About BII
About BII

The Bioinformatics Institute (BII) was set up by the Agency for Science and Technology Research (A*STAR) in July 2001; it was re-launched with a strong scientific program in the autumn months of 2007. Located in the Biopolis, BII is conceived as the computational biology research and postgraduate training institute as well as a national resource centre in bioinformatics within the Biomedical Research Council (BMRC) of A*STAR.
The BII focuses on theoretical approaches aimed at understanding biomolecular mechanisms that underlie biological phenomena, the development of computational methods to support this discovery process, and experimental verification of predicted molecular and cellular functions of genes and proteins with biochemical methods. Together with the BMRC, A*STAR research institutes and multinational R&D organizations in the Biopolis, the BII is situated in a conducive environment for exchange of scientific knowledge and friendly interaction that will prompt greater collaborations, and position the Biopolis as a notable biomedical R&D hub in Asia and in the world.

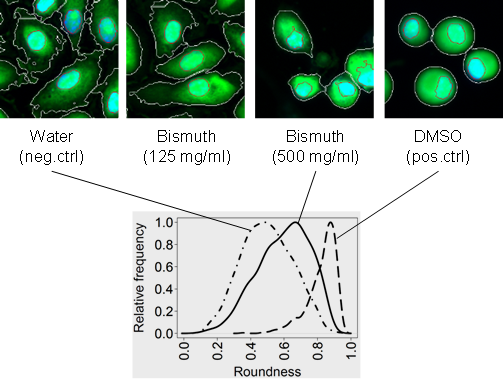 Phenotypic responses: Toxic xenobiotics often impair cellular functions and lead to changes in cellular phenotypes, such as reorganization of subcellular structures, up/down-regulation of biomolecules, or other phenotypes. Therefore, quantitative readouts based on changes in cellular phenotypes (Bougen-Zhukov et al., 2017) may be used as surrogate markers for predicting the toxicity of these chemicals. We have developed the first high-throughput and predictive in vitro nephrotoxicity assay (Loo et al., 2017a; Su et al., 2016). Our results suggest that a DNA damage response is commonly induced by different PTC toxicants that have diverse chemical structures and injury mechanisms.
Phenotypic responses: Toxic xenobiotics often impair cellular functions and lead to changes in cellular phenotypes, such as reorganization of subcellular structures, up/down-regulation of biomolecules, or other phenotypes. Therefore, quantitative readouts based on changes in cellular phenotypes (Bougen-Zhukov et al., 2017) may be used as surrogate markers for predicting the toxicity of these chemicals. We have developed the first high-throughput and predictive in vitro nephrotoxicity assay (Loo et al., 2017a; Su et al., 2016). Our results suggest that a DNA damage response is commonly induced by different PTC toxicants that have diverse chemical structures and injury mechanisms.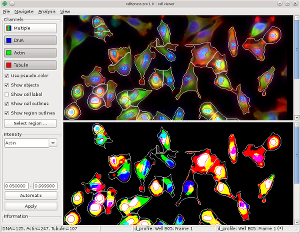 High-throughput In-vitro Phenotypic Profiling (HIPPTox): Phenotypic profiling is a computational procedure to construct quantitative and compact representations of cellular phenotypes based on the cellular images collected in high-content imaging (HCI) experiments (Bougen-Zhukov et al., 2017). We have developed several computational methods for phenotypic profiling, which include the Drug-Profiling ("D-profiling") algorithm (Loo et al., 2007) and the Protein-localization Profiling ("P-profiling") algorithm (Loo et al., 2014). We have used the phenotypic profiles constructed using these methods to classify the effects of small molecules (Loo et al., 2007, 2009), compare spatial and functional divergence of proteins (Loo et al., 2014), or predict toxicity effects of xenobiotic compounds (Su et al., 2016). The HIPPTox Platform implements many of these methods, and can be used to detect the in vitro bioactivity of chemicals and build predictive in vitro toxicity assays. The core of the platform is a user-friendly and high-performance phenotypic profiling software called "cellXpress" (
High-throughput In-vitro Phenotypic Profiling (HIPPTox): Phenotypic profiling is a computational procedure to construct quantitative and compact representations of cellular phenotypes based on the cellular images collected in high-content imaging (HCI) experiments (Bougen-Zhukov et al., 2017). We have developed several computational methods for phenotypic profiling, which include the Drug-Profiling ("D-profiling") algorithm (Loo et al., 2007) and the Protein-localization Profiling ("P-profiling") algorithm (Loo et al., 2014). We have used the phenotypic profiles constructed using these methods to classify the effects of small molecules (Loo et al., 2007, 2009), compare spatial and functional divergence of proteins (Loo et al., 2014), or predict toxicity effects of xenobiotic compounds (Su et al., 2016). The HIPPTox Platform implements many of these methods, and can be used to detect the in vitro bioactivity of chemicals and build predictive in vitro toxicity assays. The core of the platform is a user-friendly and high-performance phenotypic profiling software called "cellXpress" (