Three-dimensional Segmentation of Nuclei and mitotic Chromosomes for the study of cell divisions in live Drosophila Embryos
Rambabu Chinta and Martin Wasser

We implemented the 3D Segmentation method as a standalone application with two different graphical user interfaces (single stack segmentation and batch segmentation) using the QT user interface SDK, OpenCV (Open Computer Vision Library), the Visualization Tool Kit (VTK) and the libics v1.5 library for the Image Cytometry Standard (ICS). It supports multi-dimensional image files of the .ICS format (version 1 and 2). In single stack segmentation, the user can select an image stack from multi-time point image data, fine tune the parameters, and visualize the post-segmentation results by iso-surface viewing of segmented nuclei and displaying silhouette contours of objects on the MIP of an image stack. Segmentation parameters can be tuned using two different modes, expert or standard mode. Expert mode has 8 parameters and offers maximum control, while standard mode provides an easier to use parameter tuning. The segmentation results can be saved in three different output formats; as a statistics file that contains a list of features (e.g. ID, volume, centroid, intensity) of all 3D objects, a labeled 3D image stack in ICS format that represents the volume and surface representation in the form of sets of 2D contours. The software can be downloaded from the following download link.

GUI
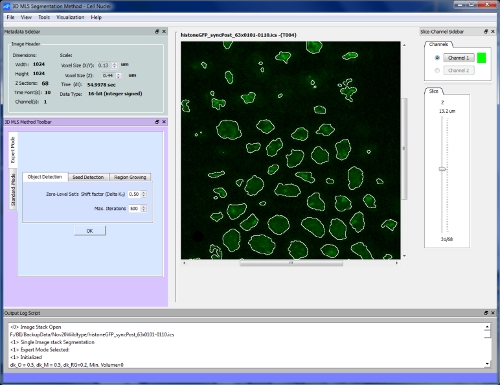

FEATURES
- File Formats
Input: Single/Multi-time point image stacks (.ics)
- Image Cytometry Standard (.ics)
Output:
- Objects features (.csv)
- Labeled 3D objects (.ics)
- ROI Contours (.cont)
- Silhouette contours(.cont)
- Image Viewing Modes
- Slices
- Max. intensity projection
- Silhouette contours
- 3D Visualization
- MIP Volume rendering
(Image stack)
- Surface rendering
(Segmented objects)
Downloads
TOOL:
- 3D MLS Cell Nuclei Detection 1.02 - Installer for all Windows plateforms (x86 and x64)
SAMPLE DATA:
- Syncytial blastoderm
- Cellular blastoderm
- Post-blastoderm (Mitotic domains)
Example

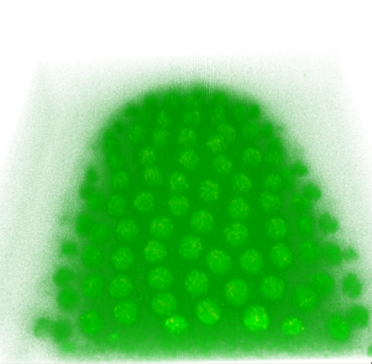
NC 12

Volume rendering of the original image stacks
Iso-surface views of segmented cell nuclei


Automatic segmentation of cell nuclei in a histone-GFP expressing live Drosophila embryo recorded at 55 Sec. intervals from Nuclear Cycle (NC) 11 until postblastoderm stage.

30 Biopolis Street, #07-01 Matrix, Singapore 138671
Copyright © 2007-2012 Bioinformatics Institute. All rights reserved.
NC 14